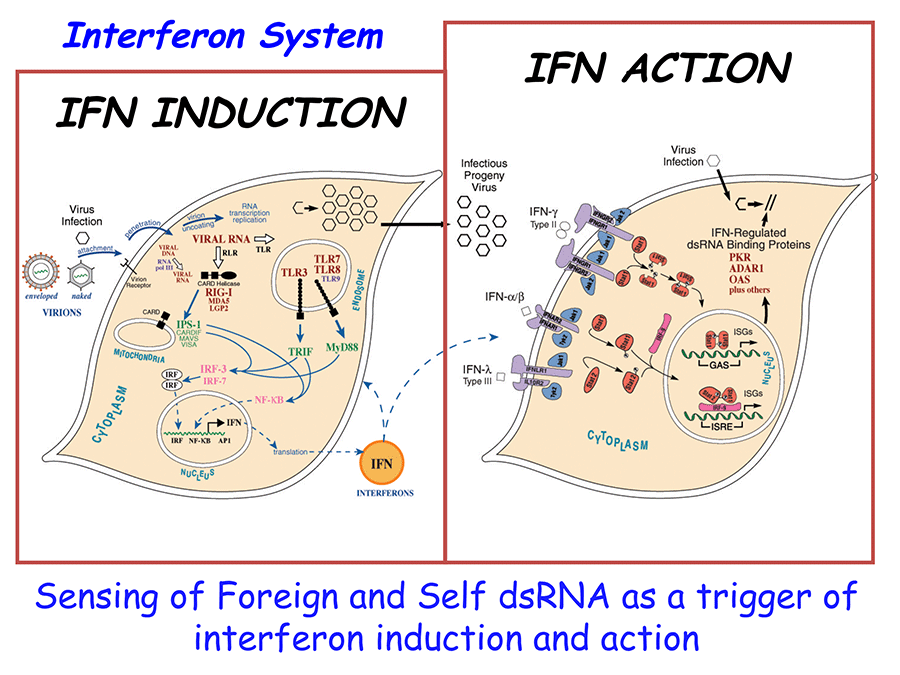
The interferon system is an important host defense system against viral and also microbiral pathogens. In addition to inhibiting virus multiplication, interferons (IFN) also affect other processes in animal cells including cell growth and differentiation and the immune response. The overall objective of our research is to elucidate in molecular terms the mechanisms by which interferons exert their antiviral and cell growth control actions in mammalian cells.
Present work in our laboratory includes the biochemical and molecular genetic study of two interferon-inducible enzymes, PKR and ADAR. PKR is an RNA-dependent protein kinase. The PKR kinase is transcriptionally induced by IFN, and is activated post-translationally by an RNA-dependent autophosphorylation. Activated PKR catalyzes the phosphorylation of the alpha subunit of protein synthesis initiation factor eIF-2. This phosphorylation causes an inhibition of protein synthesis. The PKR kinase plays a major role in the regulation of translation of viral and cellular mRNAs in animal cells. The PKR kinase is a key component of the IFN-induced antiviral response, and is responsible for the inhibition of viral protein production. PKR also acts to modulate transcription and signaling, and plays an important role in cell growth control. We are presently characterizing wild type and mutant cDNA clones of the human PKR, and the properties of animal cells transfected with these PKR cDNAs. We are also studying structures of RNAs that activate (e.g., reovirus s1 mRNA) or antagonize activation (e.g., adenovirus VA RNA and HIV TAR RNA) of the human kinase. Finally, we have defined the intron-exon organization and promoter structure of the human and mouse PKR genes, and are characterizing the signal transduction leading to activation of PKR transcription.
As part of our interferon studies, we have isolated and characterized cDNA and genomic clones of the IFN-regulated human adenosine deaminase, ADAR1. ADAR1 deaminase is an RNA editing-enzyme, which catalyzes the C-6 deamination of adenosine to yield inosine. The editing of the cellular pre mRNAs for glutamate and serotonin neurotransmitter receptors and the editing of viral RNA genomes during lytic and persistent infections is catalyzed by ADAR. We are now using a combined molecular and cell biological approach to study the structure, function, and expression of the interferon-regulated adenosine deaminase, and the role of the deaminase in viral pathogenesis.
In studies complementary to those on IFN action, we are also investigating the molecular basis of translational control in the reovirus system and, with investigators in the Materials Research Laboratory, the development of cationic liposome systems for efficient gene delivery. Our reovirus studies are focused on the expression of the s-class mRNAs and the functional properties of their encoded proteins. The s1 mRNA is bicistronic, encoding the virus attachment receptor protein. The s3 and s4 mRNAs both encode RNA binding proteins, one of which antagonizes the function of the PKR kinase.