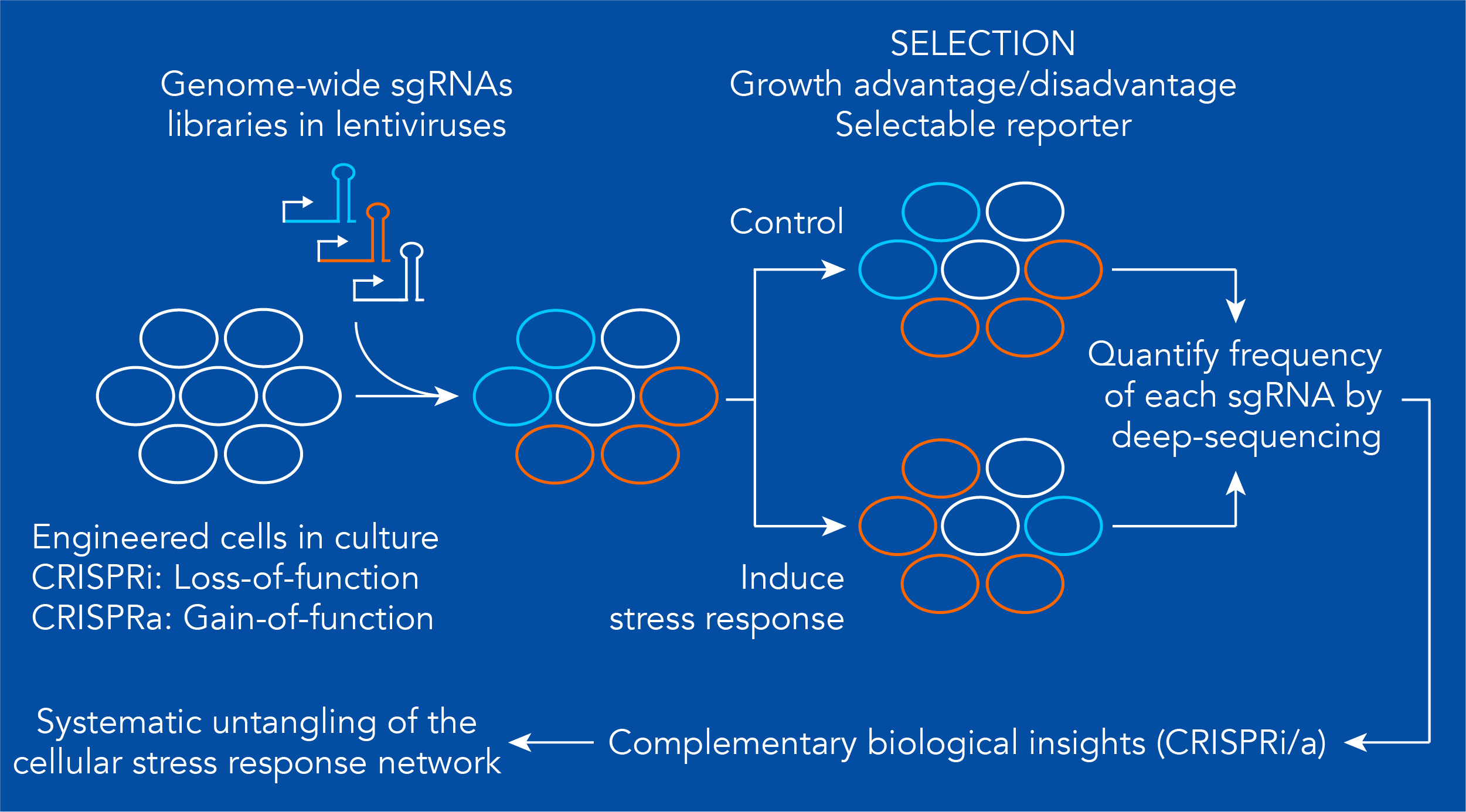
Genetic modulators of stress responses
The output of any cellular stress response depends on the intensity and duration of the stress signal and the concerted actions of multiple genes activated in response to the stress signal. These genes can amplify or dampen the output of specific signaling pathways, thereby fine-tuning the stress response. Parallel CRISPR-based gain- and loss-of-function genetic screens based on modifications of the CRISPR/Cas9 genome editing technology have the power to identify the genetic modulators of the unfolded protein response (UPR) and the RNA stress response (RSR). In these screens, “designer transcription factors” that work by fusing a catalytically dead Cas9 nuclease to transcription repressor (CRISPRi) or transcription activator (CRISPRa) domains are employed to silence or turn on genes, respectively. Genome-wide libraries of small guide RNAs (sgRNAs) are then introduced into cells that express these Cas9 fusions (using a single sgRNA per cell). Subsequently, the cells are exposed to unfolded protein or harmful RNA stress, or to the forced-activation of key UPR or RSR components, and screened for a selectable phenotype, such as changes in proliferation or survival, or the induction of fluorescent reporters that allow collecting cells by flow cytometry. The frequency of each sgRNA in the cell populations is determined by deep sequencing. This screening platform allows pinpointing the genes that modulate each stress response. We have employed a similar shRNA-based screening platform to identify the genes that control the response and adaptation of cancer cells to proteasome inhibitors (Acosta-Alvear, et al., eLife 2015).
A major consequence of inducing the UPR or the RSR is the global attenuation of translation resulting from activation of PERK or PKR, respectively, and phosphorylation of eIF2α downstream of either. This signaling cascade leads to the preferential translation of select mRNAs, such as those containing regulatory uORFs (see “How do cells sense, respond, and adapt to protein folding stress or harmful RNAs?” for details). The resulting changes in abundance of specific proteins could add another layer of control to fine-tune the UPR or the RSR. Ribosome profiling, a well-established deep sequencing based method that allows the transcriptome-wide measurement of ribosome loading on mRNAs, can yield valuable insights into the translation of all transcripts in cells experiencing protein-folding stress or harmful RNA-induced stress.





